Last December as I was preparing to teach a night class on compilers and programming languages, I looked for a poster of programming languages to hang in the classroom to give students (and myself) a sense of how many programming languages we didn’t know and how the unknown languages related to each other and languages we did know.

I figured I was looking for a tree of programming languages, thinking of the way an evolutionary tree shows that birds are more related to lizards than they are mammals. After reading The Tangled Tree by David Quammen this summer I’ve been able to unpack my implicit assumption that I could usefully organize programming languages in a tree like this. I’m drawing liberally from that text here, so if anything here sounds smart you should read the book to get the full heft of it.
Since my goal was to provide a survey of programming languages (without, unfortunately, dedicating almost any class time to this; it would have been fairer to call the course “Let’s mostly learn about the compilation and interpretation of Python and also write our own interpreters”), I think what I really wanted was a visualization of what the languages were like, not where there came from. These kinds of trees have existed for plants and animals for a long time: although Aristotle used a linear hierarchy to distinguish rocks from plants from animals from humans from angels from God, for the last few hundred years a branching tree has been a natural way to classify fractally diverse domains like “what animals are there and which should we think about together.”

Putting, say, marine animals on one branch, land animals on a second, and flying animals on a third, needn’t originally mean that all flying animals were more genetically or ancestrally related to each other than any one of the was to a land animal.t and then by color, we are portrayed these distinctions as than the fundamental difference of land, sea, or air. While this representations encodes that the ability to fly is a more fundamental distinction than being a bird or not, it says nothing – at least intentionally – about the historical record of evolution of these animals or the amount of genetic material they share. Modern field guide keys that present decision trees are similar: first, are you in North America? Second, is the bird black? Third, is it larger than a breadbox?

I had previously found a grid of functional vs object oriented languages and dynamically typed vs statically typed languages on two axes useful when defining these terms for students:
dynamically typed statically typed
functional Erlang Haskell
Elixir ML
Scheme minus set! Elm
imperative Python C
JavaScript Java
Ruby
This could be turned into a tree by choosing one of these characteristics as more fundamental or more distinguishing to put closer to the root; I’d choose function vs imperative as more fundamental.
Elixir
Erlang| | /
\ /_/ /
\Scheme/ | / ML
\ / / / Haskell | | |Ruby| /C/
\ | / / | |/ | |Python| | | / / /Java/
\ \/ \ | | | \/ / / / / /
\ \ | statically/ | / / |___/ /
\dynamically\ | typed / | dynamically/ / /
\ typed \| / | typed / /statically/
\ / | / / typed /
\ / \ /_/ /
\ / \ /
\ | | ______/
\ \ / /
\ functional \ / imperative/
\ \ / /
\ \ / /
\ \__/ /
\ /
| |
| |
| programming languages |
| |
| |
Again, in this sort of tree position says nothing about history. This tree is information technology for displaying diversity.
Looking for this a tree of programming languages was deep rabbit hole, made especially attractive and dangerous by needing to finish the curriculum I’d be teaching my vacation was over. After a few hours I’d decided I definitely wasn’t going to make my own chart, and I found this this old poster from O’Reilly:

It goes the more some nice properties: specific releases of programming languages are shown at points in time, according to label along the x axis; at the births of languages, arrows indicate other languages that influenced that one; and it’s quite visually appealing. History can be a useful way to frame concepts, and I could imagine students tracing their programming languages of choice through its evolution and tracing back to others that inspired it. And some bummer properties: the timeline ends in 2004; the chart is really big; and it’s hard to know how the ancestor languages were chosen.

This sort of tree has been around a long time too: before Darwin published “On the Origin of Species,” trees depicting the geologic record showed the introduction of new species (presumably by a God with a Plan) and the extinction of others.
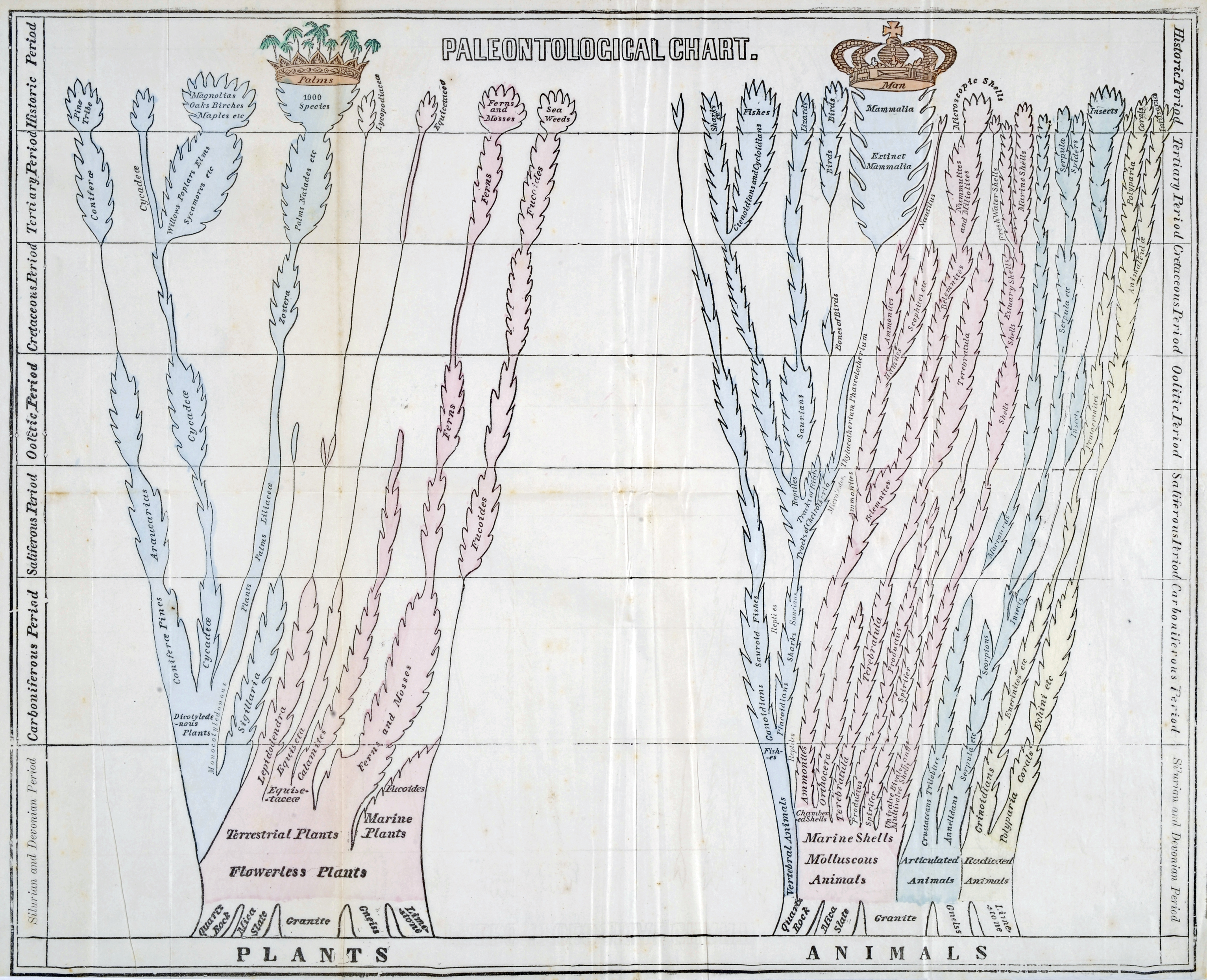
Darwin originally used the metafore of tree coral for his branching species of mockingbirds, since only the tips of a tree coral are alive — matching the way the ancestral tree of life grows only at the sprouting branches, the rest of the tree being composed of now-extinct common ancestor species. While less accurate in this respect, perhaps switching to a tree helped his hypothesis loom so large in minds at the time because it co-opted an existing metafore: those trees you’re already using to describe the diversity of life? There’s another dimension already depicted: those branches are speciation, and it’s happening all the time!
If we define programming languages as still alive meaning they are still in common use, then the ancestor-still-being-alive aspect of a tree works well for us classifying programming languages.
We have our own fossil record for creating these trees, a resource geneticists could only dream of: we read the timestamps in their notebooks and commit messages of the code artifacts that were the release of a programming language. The historical record of the actual artifacts — the code for the implementation of these languages — is often clear. We have journals and magazines with advertisements for new versions of BASIC.
Eventually, evidence from molecular biology made constructing these biologic phylogenetic trees a more computational task: the molecular clock of genetic drift as species diverged made estimating the time since divergence a complex but quantitative matter of comparing sequences of DNA. This new information shook up and rearranged a lot of existing trees! But it was a solid, consistent technique, useful not only for dating evolutionary branching events but also for a measure of similarity.
The links between programming languages on the graph seem useful, but they’re suspect. There’s a project to indicate each of these types of relationships and to excise those that can’t be backed up.
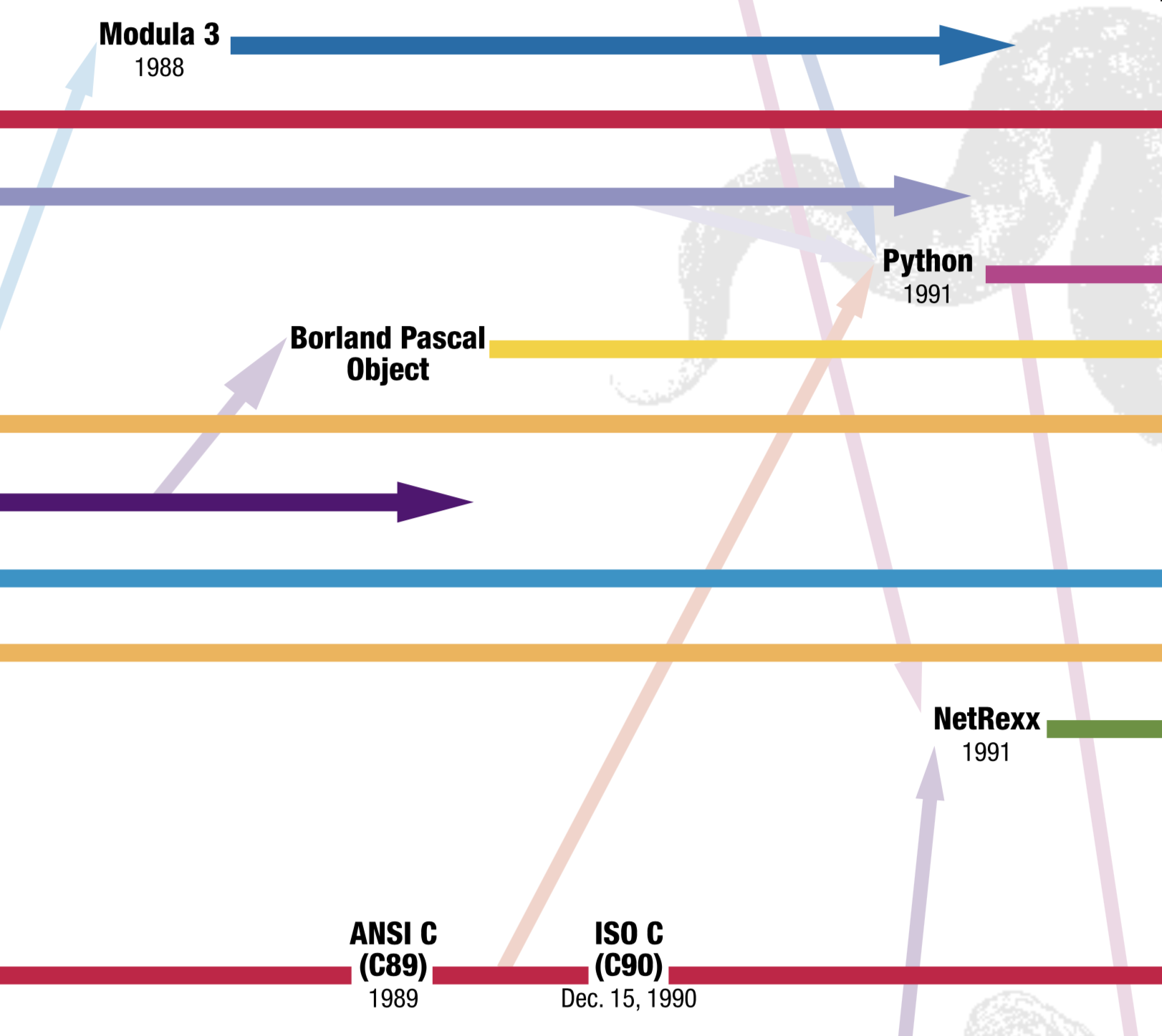
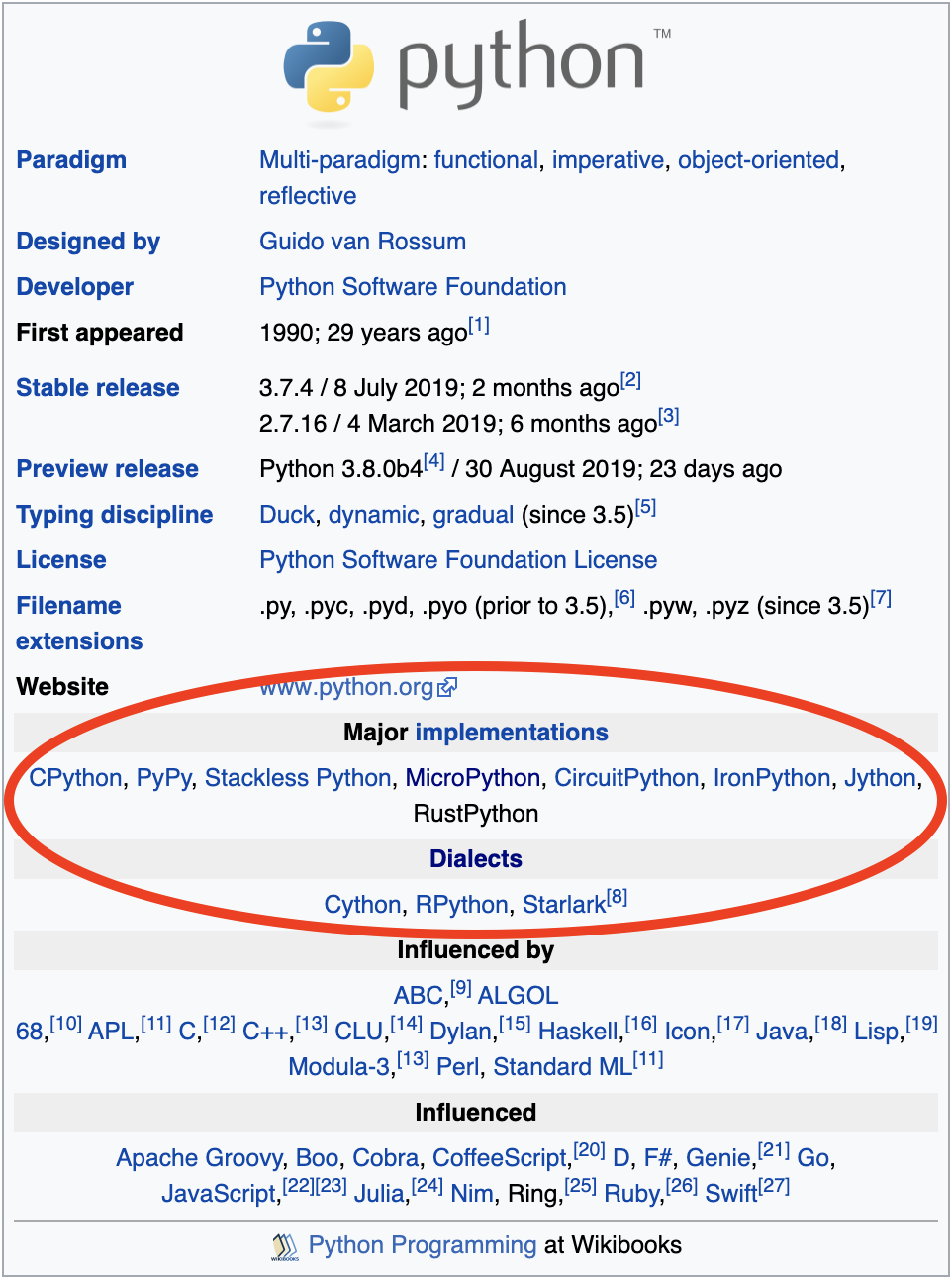
The chart I found depicts Python as inspired from C and Modula 3. Wikipedia cites 12 more languages too.
TOM TODO HERE rewrite below this line - this is too dressed up.
It’s temping to look for a DNA analogue for our programming languages, source code being the natural thing to jump to; changes to a given language implementation indeed have some correspondence to the scope of those changes — but that underestimates how similar e.g. cleanroom implementations of a language are. Building phylogenetic trees with source code is like trying to measure the relatedness of the political platforms of two candidates by doing plagiarism detection on their speeches. The actual text of stump speeches may be too granular such that two very similar platforms - even candidates who might support the same legislation - might not contain the same phrases. It’s just too common to sit down to an empty text editor and write a new language, drawing ideas from existing languages but not inheriting the cruft of an older codebase.
Sounds like a job for memetics, Dawkins’ “ideas behave like genes, sort of, well a lot actually” big idea. Which syntactical constructs are reused? Which proramming languages features? Take all the big words you could use to describe a language - how many of these words apply to another language? But then we’re back to making field guides.
We have another tool - we can interview the language creators themselves and ask what their inspirations were!
TODO: add exerpt from History of Programming Languages
Seeing the video is a simple matter of breaking it out of the ACM’s vault.
As we track ideas between programming languages another correspondence to biological phylogenetic trees pops out. Studying bacteria in particular, a tremendous amount of “horizontal gene transfer” has been observed: in construct phylogenetic trees with genetics, it’s sometimes necessary to show branches converging when genes present in two species are not present in their shared ancestral species: when species pass genes to one another.
It’d be nice to draw these in too! Clearly Python 3.7 followed from Python 3.6, but a bit of horizontal transfer from JavaScript exists for the async and await keywords. EcmaScript 6 took generators (the yield keyword) from Python. List comprehensions came from Haskell! The same way the bacterial world turned out to share genes not just with offspring, ideas in programming languages can be shared without sharing a common ancestor with that feature.
TODO show a reticulated tree!
“On the Origin of Species” was about, well, species. Biological species are intuitively lifeforms that feel really similar to each other, and technically lifeforms that can mate and produce viable offspring.
The abundance of horizontal transfer brings into question our notion of species.
Darwin was dealing with the concept of speciation - how new species emerge from old? (his answer: separation + random mutations * time leads to inability to reproduce fertile offspring).
For programming languages, a dialect might be different if there are programs which run in one dialect but not another - maybe the portion of relation might be the portion of programs which work in both languages?
The SQL family of languages mostly implement an official spec, then add their own special features on top.
The Python ecosystem has a variety of implementations with different behavior. Although most of these would be considered “not technically Python” since they do not implement the specification (a veritable definition, “if you are implementing Python, this code must behave in this way”) some relative similarity is worth measuring.
A recent call for new implementations of Python that are faster or run on new platforms makes this speciation concept timely to examine. Will the next implementation really be Python? Is it something else?
the Python ecosystem is about to become more crowded.
For the domain of life that seems to share genes as freely as ideas in programming languages seem to hop around, an idea was presented that we might consider all bacteria on earth to be a single species — even a single organism! – because once a trait is found by individual it can spread so freely.
This seems like a bit of a cop out for our definition: we want to be able to say whether something is Python.
Looking to other languages: is such and SQL? And not “compliant” SQL, but is it SQL? Or is it “like SQL?”
Noting differences between languages and measuring degree of similarity is tremendously important for pedagogy: how hear learn and teach a programming languages. And inthe Python world, we are on the cusp of a bunch of learning that will need to be done: a call has gone out for a new Python implementation, and this new implementation might not - will almost certainly not - be completely compatible with CPython.
At this point in writing this I get that shiver on the back of my spine that I’m about to recreate a field of study I know nothing about in the most naive, broken way - in this case, comparative linguistics.
xkcd of not understanding a topic
So I’ll back away from this comparison. It may be that the analogy of programming languages to natural languages would bear more fruit here, but I’ll stick with comparing programming language implementations - artifacts of code - to biological organisms.
What’s the different between a tree and a web, in the data structures?
What makes a species distinct? What makes a species viable?
Speciation - python implementations - talk about this in another post.
Thinking about new Python languages.
Preparing for a language I don’t know about yet